mop calculates a Mobility-Oriented Parity
mop(
M_calibra,
G_transfer,
percent = 10,
comp_each = NULL,
normalized = TRUE,
standardize_vars = TRUE
)Arguments
- M_calibra
A SpatRaster of variables representing the calibration area (M area in ENM context).
- G_transfer
A SpatRaster of variables representing areas or scenarios to which models will be transferred.
- percent
(numeric) percent of values sampled from the calibration region to calculate the MOP.
- comp_each
(numeric) compute distance matrix for a each fixed number of rows (default = 100). This parameter is for paralleling the MOP computations using the furrr package, see example 2.
- normalized
(logical) if TRUE mop output will be normalized to 1.
- standardize_vars
Logical. Standardize predictors see
std_vars
Value
A SpatRaster object of MOP distances.
Details
The MOP is calculated following Owens et al. (2013; doi:10.1016/j.ecolmodel.2013.04.011 ). This function is a modification of the MOP function in ntbox R package.
Examples
# Predictors
m_path <- system.file("extdata/M_layers", package = "smop") |>
list.files(full.names=TRUE)
g_path <- system.file("extdata/G_layers", package = "smop") |>
list.files(full.names=TRUE)
M_calibra <- terra::rast(m_path)
G_transfer <- terra::rast(g_path)
# Example 1 compute mop distance in serial
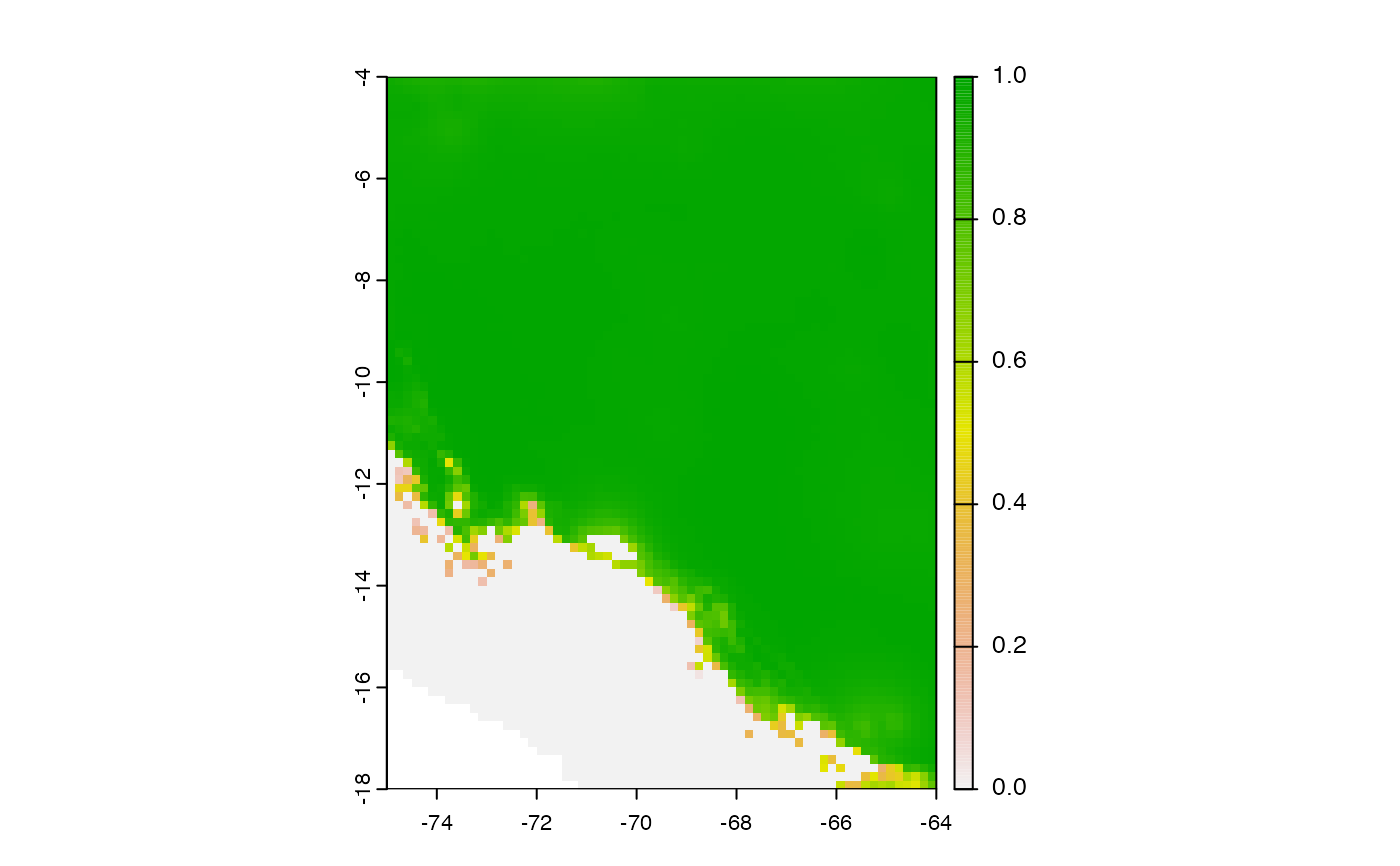
mop_test <- smop::mop(M_calibra = M_calibra, G_transfer = G_transfer,
percent = 20,comp_each = NULL,
normalized = TRUE, standardize_vars=TRUE)
terra::plot(mop_test)
 # \donttest{
# Example 2: Run the mop function in parallel
# future::plan("future::multisession",workers = 2)
# mop_test_parallel <- smop::mop(M_calibra = M_calibra,
# G_transfer = G_transfer,
# percent = 20,comp_each = 500,
# normalized = TRUE, standardize_vars=TRUE)
# future::plan("future::sequential")
# terra::plot(mop_test_parallel)
# }
# \donttest{
# Example 2: Run the mop function in parallel
# future::plan("future::multisession",workers = 2)
# mop_test_parallel <- smop::mop(M_calibra = M_calibra,
# G_transfer = G_transfer,
# percent = 20,comp_each = 500,
# normalized = TRUE, standardize_vars=TRUE)
# future::plan("future::sequential")
# terra::plot(mop_test_parallel)
# }