bam_sim: Simulate dispersal dynamics using the set B of the BAM framework.
Source:R/bam_sim.R
bam_sim.Rdbam_sim: Simulate dispersal dynamics using the set B of the BAM framework.
Usage
bam_sim(
sp1,
sp2,
set_M,
initial_points,
periods_toxic,
periods_suitable,
nsteps,
progress_bar = TRUE
)Arguments
- sp1
Niche model of the focal species (the one that disperses).
- sp2
Niche model of the species with whom sp1 interacts (currently no dispersal dynamics for this species).
- set_M
A setM object containing the adjacency matrix for sp1. See
adj_mat- initial_points
A sparse vector returned by the function
occs2sparse- periods_toxic
Time periods that sps2 takes to develop defense mechanisms (i.e. toxic).
- periods_suitable
This is the time that sp2 takes to become non-toxic
- nsteps
Number of steps to run the simulation
- progress_bar
Show progress bar
Value
An object of class bam. The object contains 12 slots of information (see details) from which simulation results are stored in sdm_sim object, a list of sparse matrices with results of each simulation step.
Details
The returned object inherits from setA,
setM classes. Details about the dynamic model
can be found in Soberon and Osorio-Olvera (2022). The model is cellular
automata where the occupied area of a species at time \(t+1\) is
estimated by the multiplication of three
binary matrices: one matrix represents movements (M), another
abiotic -niche- tolerances (A), and a third, biotic interactions (B)
(Soberon and Osorio-Olvera, 2022).
$$\mathbf{G}_j(t+1) =\mathbf{B}_j(t)\mathbf{A}_j(t)\mathbf{M}_j
\mathbf{G}_j(t)$$
References
Soberón J, Osorio-Olvera L (2023). “A dynamic theory of the area of distribution.” Journal of Biogeography6, 50, 1037-1048. doi:10.1111/jbi.14587 , https://onlinelibrary.wiley.com/doi/abs/10.1111/jbi.14587. .
Examples
# Compute dispersal dynamics of Urania boisduvalii as a function of
# palatable Omphalea
urap <- system.file("extdata/urania_omph/urania_guanahacabibes.tif",
package = "bamm")
ura <- raster::raster(urap)
ompp <- system.file("extdata/urania_omph/omphalea_guanahacabibes.tif",
package = "bamm")
omp <- raster::raster(ompp)
msparse <- bamm::model2sparse(ura)
init_coordsdf <- data.frame(x=-84.38751, y= 22.02932)
initial_points <- bamm::occs2sparse(modelsparse = msparse,init_coordsdf)
set_M <- bamm::adj_mat(modelsparse = msparse,ngbs = 1)
ura_sim <- bamm::bam_sim(sp1=ura, sp2=omp, set_M=set_M,
initial_points=initial_points,
periods_toxic=5,
periods_suitable=1,
nsteps=40)
#>
|
| | 0%
|
|== | 2%
|
|==== | 5%
|
|===== | 8%
|
|======= | 10%
|
|========= | 12%
|
|========== | 15%
|
|============ | 18%
|
|============== | 20%
|
|================ | 22%
|
|================== | 25%
|
|=================== | 28%
|
|===================== | 30%
|
|======================= | 32%
|
|======================== | 35%
|
|========================== | 38%
|
|============================ | 40%
|
|============================== | 42%
|
|================================ | 45%
|
|================================= | 48%
|
|=================================== | 50%
|
|===================================== | 52%
|
|====================================== | 55%
|
|======================================== | 58%
|
|========================================== | 60%
|
|============================================ | 62%
|
|============================================== | 65%
|
|=============================================== | 68%
|
|================================================= | 70%
|
|=================================================== | 72%
|
|==================================================== | 75%
|
|====================================================== | 78%
|
|======================================================== | 80%
|
|========================================================== | 82%
|
|============================================================ | 85%
|
|============================================================= | 88%
|
|=============================================================== | 90%
|
|================================================================= | 92%
|
|================================================================== | 95%
|
|==================================================================== | 98%
|
|======================================================================| 100%
ura_omp <- bamm::sim2Raster(ura_sim)
#>
|
| | 0%
|
|== | 2%
|
|==== | 5%
|
|===== | 8%
|
|======= | 10%
|
|========= | 12%
|
|========== | 15%
|
|============ | 18%
|
|============== | 20%
|
|================ | 22%
|
|================== | 25%
|
|=================== | 28%
|
|===================== | 30%
|
|======================= | 32%
|
|======================== | 35%
|
|========================== | 38%
|
|============================ | 40%
|
|============================== | 42%
|
|================================ | 45%
|
|================================= | 48%
|
|=================================== | 50%
|
|===================================== | 52%
|
|====================================== | 55%
|
|======================================== | 58%
|
|========================================== | 60%
|
|============================================ | 62%
|
|============================================== | 65%
|
|=============================================== | 68%
|
|================================================= | 70%
|
|=================================================== | 72%
|
|==================================================== | 75%
|
|====================================================== | 78%
|
|======================================================== | 80%
|
|========================================================== | 82%
|
|============================================================ | 85%
|
|============================================================= | 88%
|
|=============================================================== | 90%
|
|================================================================= | 92%
|
|================================================================== | 95%
|
|==================================================================== | 98%
|
|======================================================================| 100%
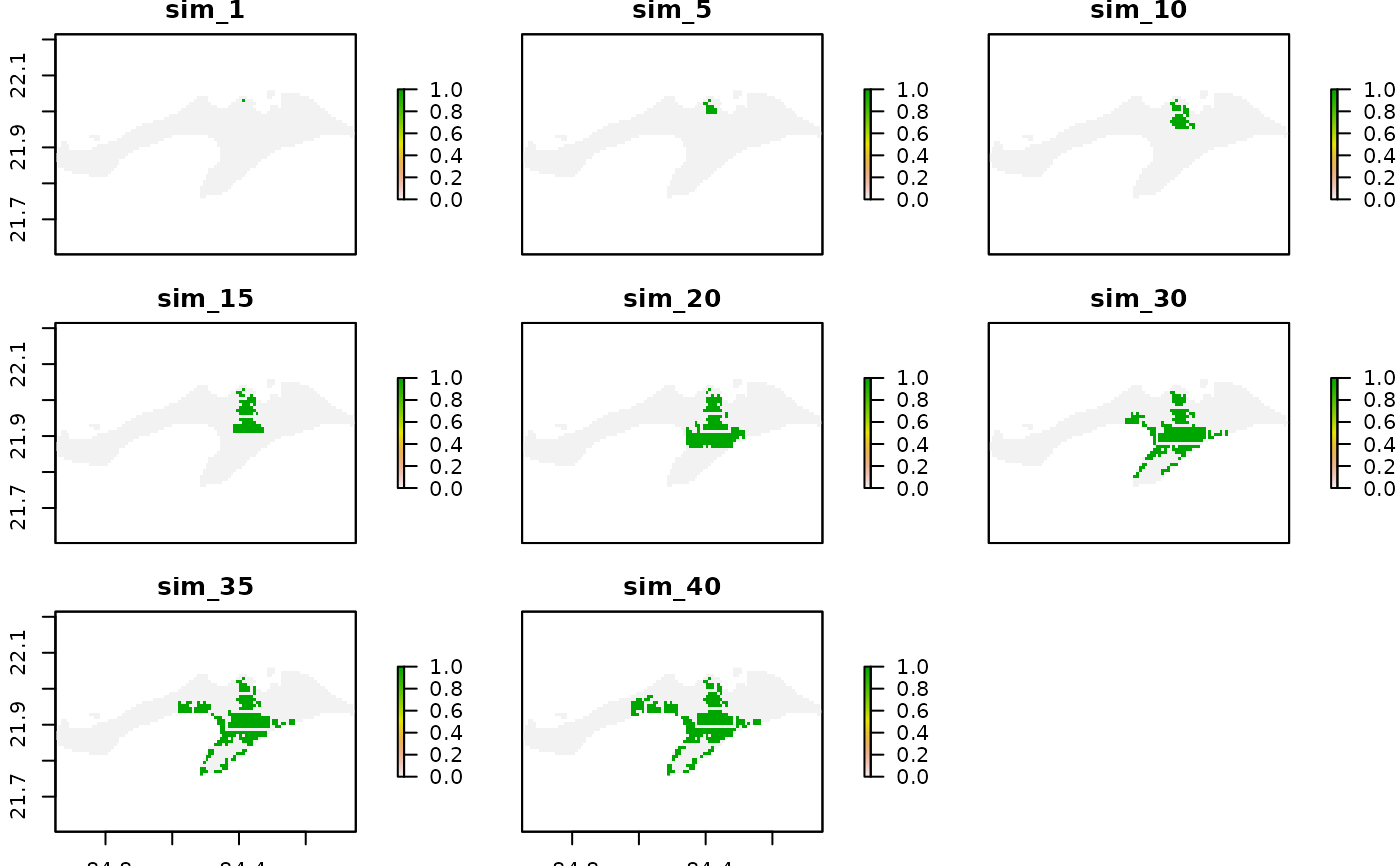
raster::plot(ura_omp[[c(1,5,10,15,20,30,35,40)]])
 # \donttest{
if(requireNamespace("animation")){
# Animation example
anp <-tempfile(pattern = "simulation_results_",fileext = ".gif")
# new_sim <- bamm::sim2Animation(sdm_simul = ura_sim,
# which_steps = seq_len(ura_sim@sim_steps),
# fmt = "GIF",
# filename = anp)
}
#> Loading required namespace: animation
# }
# \donttest{
if(requireNamespace("animation")){
# Animation example
anp <-tempfile(pattern = "simulation_results_",fileext = ".gif")
# new_sim <- bamm::sim2Animation(sdm_simul = ura_sim,
# which_steps = seq_len(ura_sim@sim_steps),
# fmt = "GIF",
# filename = anp)
}
#> Loading required namespace: animation
# }